Data Files (JSON and RDF)
A collection of flat files representing the bulk of the PACER case data that SCALES has purchased over its lifetime, available here for users with Satyrn credentials.
We offer two file formats for the case files themselves: JSON (JavaScript Object Notation) and Turtle (Terse RDF Triple Language). The JSON files are parsed versions of HTML files downloaded from PACER. The Turtle files are the product of a SCALES script that (1) straightforwardly translates the JSON files into a graph-data paradigm and (2) pulls in additional data sources that we deemed research-worthy: our Litigation Ontology labels, our Entity Disambiguation dataset, and the Federal Judicial Center's Integrated Database (IDB) and judge biography directory.
For both file types, the files have been sorted into subdirectories based on year code (a PACER case-id convention that may not correspond to the actual filing year) and court. Furthermore, in the uppermost Turtle directory, a file called courts.ttl contains some case-nonspecific triples relating to the federal courts.
Case lists
For convenience, alongside the case files hosted on our file server, we also maintain a folder of CSV case-number lists describing the various categories of data we possess (some of which exceed the scope of the case files on the server). Many of the lists enumerate the results of scraper runs chronicled in the pipeline documentation. The categories are defined as follows:
- 2016: cross-sectional data for '16, non-N.D.-Ill (247,416 cases)
- 2017: cross-sectional data for '17, non-N.D.-Ill (319,718 cases)
- clearinghouse: misc AL/GA/IL data scraped for Civil Rights Litigation Clearinghouse (10,483 cases)
- cross_sectional: cases in the cross-sectional dataset on the file server (676,809 cases)
- gand: longitudinal data in N.D. Ga, 2010 & 2015 (11,398 cases)
- ilnd: longitudinal data in N.D. Ill, 2002 – Mar '21 (199,664 cases)
- longitudinal: cases in the longitudinal dataset on the file server, same as above (199,664 cases)
- mistakes: cases scraped accidentally, usually because of PACER filing-date ambiguity (275 cases)
- mongo: cases available in our Mongo database (879,842 cases)
- nacdl: partially cross-sectional NoS-440 case stubs scraped for NACDL (4694 cases)
- paed: longitudinal data in E.D. Pa for private use only, 2002-21 (326,102 cases)
- recap: cases obtained from the Free Law Project's RECAP archive (86,169 cases)
- satyrn: cases available for analysis in the Satyrn app (864,700 cases)
- tx: longitudinal data in Texas, 2018 & 2020 (72,349 cases)
- all (1,278,268 cases)
Schemas
Because the JSON files come directly from our PACER parser, the JSON Schema section of the parser documentation specifies their structure.
The following flowchart specifies the resources and properties described in the Turtle files (each node-edge-node relation in the chart represents a class of "resource -> property -> resource" triples that can appear in the files):
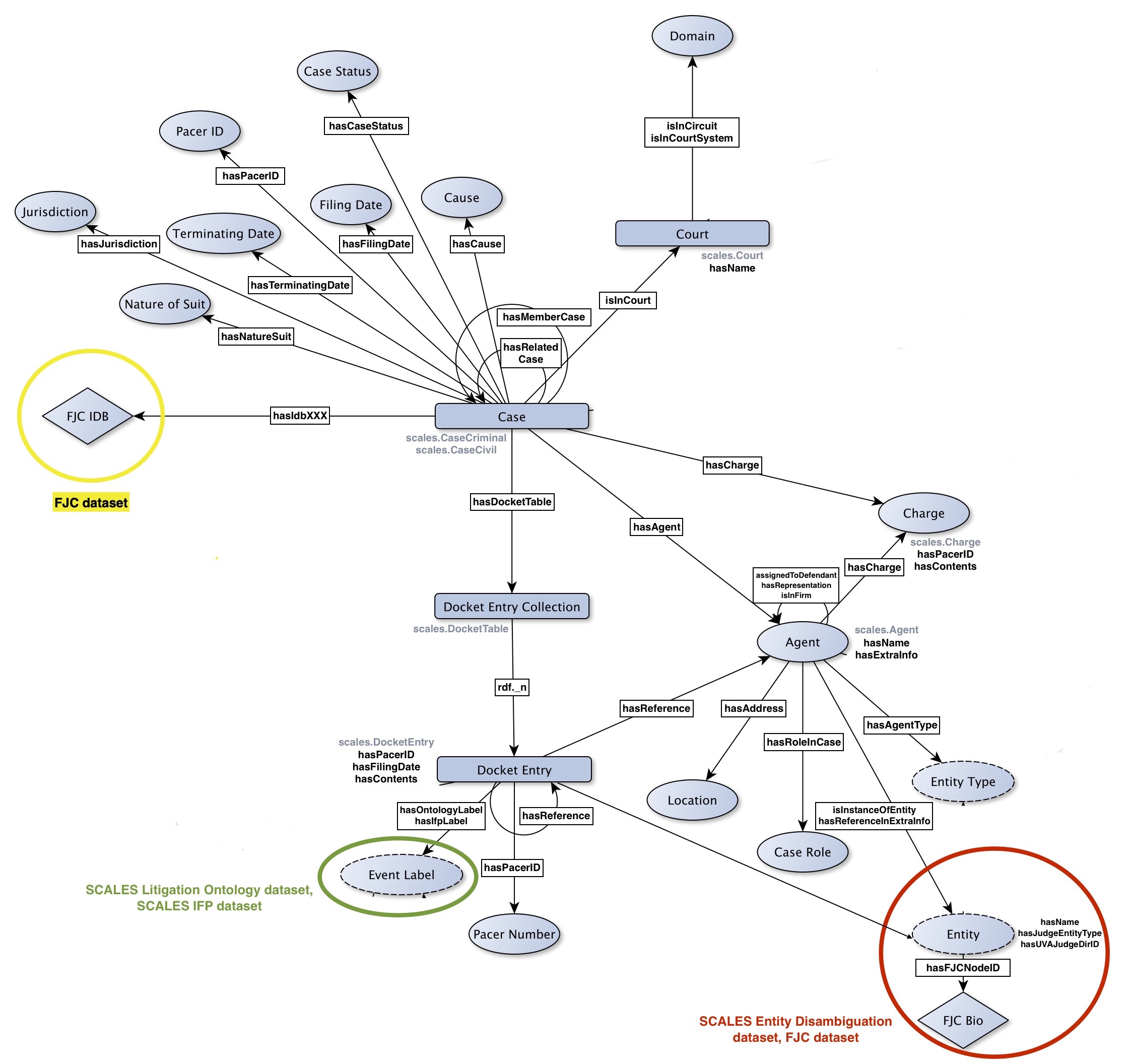
A few notes:
- For visual clarity, a few predicates (e.g. "hasName" under "scales.Court") are represented by their name alone, rather than a labeled edge to a node; there is otherwise no particular significance to these predicates.
- The edge labeled "hasIdbXXX" refers to a family of predicates named after IDB columns ("hasIdbDocket," "hasIdbFiledate," etc). For a given case subject, these predicates point to literals copied from the IDB row corresponding to that case.
- The edge labeled "rdf._n" denotes predicates of the form "rdf._0,", "rdf._1," etc, which follow an RDF convention dictating that the objects to which such predicates point can be considered items in an ordered list.
- For a variety of reasons (null values, features of criminal cases that don't appear in civil cases or vice versa, and so on), each individual case file will include only a subset of the possible triple types described above.
Usage examples
Iterative loads of the JSON files are usually quite fast, which makes them a versatile first-line research solution, especially when you want to do things like assemble dataframes or operate on strings. For instance, the code block below uses a simple string comparison to flag corporate litigation that may involve Apple:
import json
import glob
matched_paths = set()
for json_path in tqdm(glob.glob('path/to/files/scales_json_cross_sectional/*/*/*.json')):
json_data = json.load(open(json_path))
for party in json_data['parties']:
if party['name'] is not None and 'Apple' in party['name']:
matched_paths.add(json_path)
On the other hand, the Turtle files are much slower to load into working memory — but what they lack in lightweightness they make up in structure, particularly when it comes to relationships between entities. Also, the inclusion of non-PACER datasources allows for a wider variety of inquiries, like the below code that searches the previously identified cases for agents with the exact name "Apple Inc." and ascertains those agents' identities as disambiguated by SCALES:
import rdflib
from rdflib import Graph
scales = rdflib.Namespace('http://schemas.scales-okn.org/rdf/scales#')
g = Graph()
for json_path in tqdm(matched_paths):
g.parse(json_path.replace('scales_json','scales').replace('.json','.ttl'))
agent_iris, entity_iris = set(), set()
# see rdflib.readthedocs.io/en/stable/intro_to_graphs.html
for s,p,o in g.triples((None, scales.hasName, rdflib.Literal('Apple Inc.'))):
agent_iris.add(s)
for iri in agent_iris:
for s,p,o in g.triples((s, scales.isInstanceOfEntity, None)):
entity_iris.add(o)
print(entity_iris) # it turns out this is a 1-element set, as all the agents we found are instances of the same entity
Since Turtle is a syntax for expressing the versatile RDF data model, these files are well-suited to many languages and conventions. Even within Python there are a number of interaction options, such as the subjects(), objects(), and predicates() methods on rdflib Graph objects:
apple_iri = list(entity_iris)[0]
apple_aliases, apple_lawyers = set(), set()
for s,p,o in g.triples((None, scales.isInstanceOfEntity, apple_iri)):
for name in g.objects(s, scales.hasName):
apple_aliases.add(str(name))
for lawyer_iri in g.objects(s, scales.hasRepresentation):
apple_lawyers.add(lawyer_iri)
print('Found', len(apple_aliases), 'aliases:', apple_aliases)
print('Found', len(apple_lawyers), 'lawyers')
If you prefer, you can query the Turtle files using SPARQL, which enables complex constructs like the below query identifying disambiguated pairs of lawyers who represented Apple together:
query = f"""
SELECT DISTINCT ?iri1 ?iri2 WHERE {{
?party scales:isInstanceOfEntity <{apple_iri}> .
?party scales:hasRepresentation ?lawyer1 .
?party scales:hasRepresentation ?lawyer2 .
?lawyer1 scales:isInstanceOfEntity ?iri1 .
?lawyer2 scales:isInstanceOfEntity ?iri2 . FILTER (?iri1 != ?iri2)
}}"""
for row in g.query(query):
print(f"{row.iri1} appears with {row.iri2}") # t
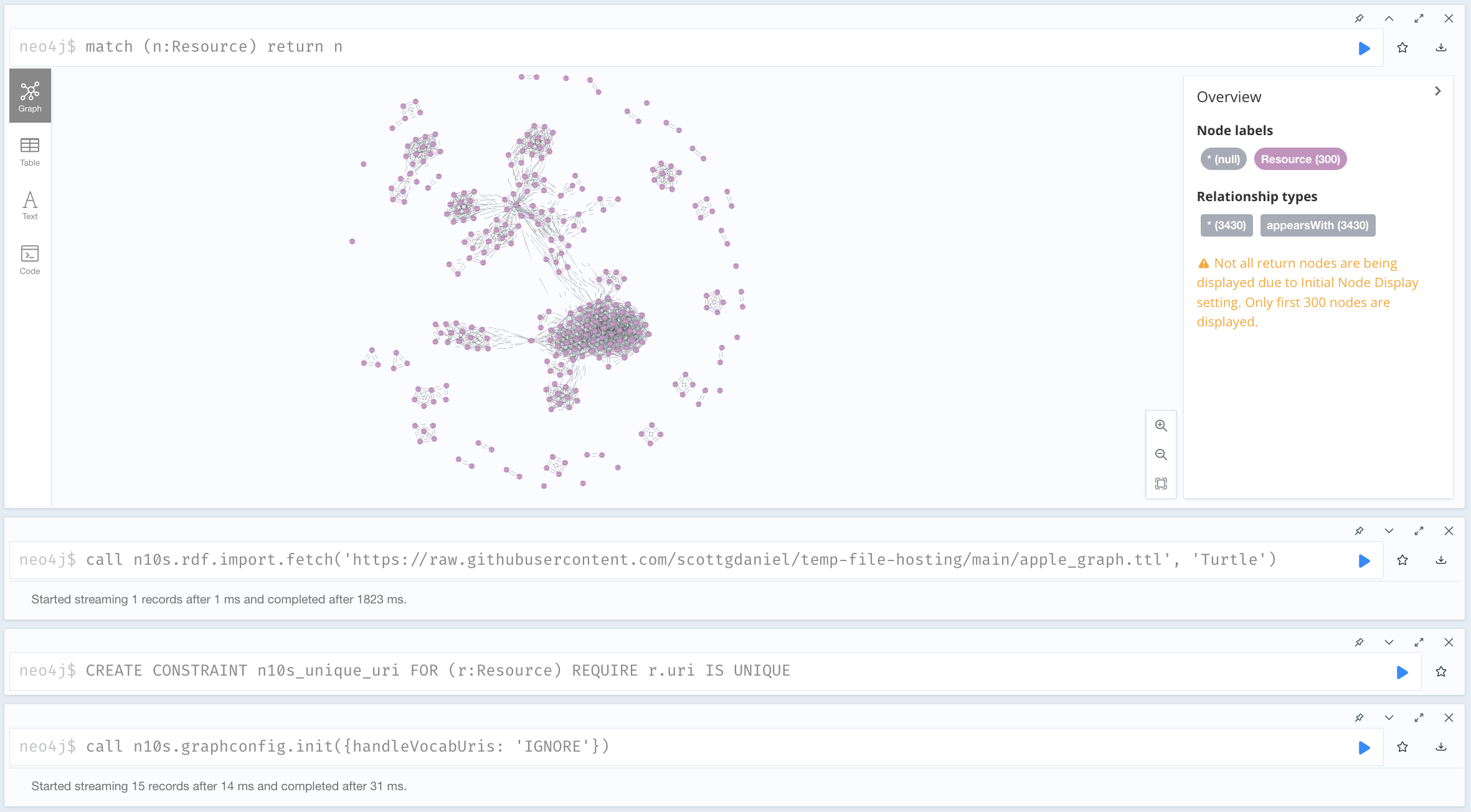
You can even create visualizations of your results. For example, it's easy to export the above query to a file...
graph_for_export = Graph()
for row in g.query(query):
graph_for_export.add((row.iri1, scales.appearsWith, row.iri2)) # new predicate, for example purposes
graph_format = 'ttl'
with open(f'apple_graph.{graph_format}', 'w') as f:
f.write(graph_for_export.serialize(format=graph_format))
...at which point it can be imported into a graph-visualization platform like Neo4j Bloom:
Feel free to get in touch to request access to our data files if these examples piqued your interest, and/or register for our research tool Satyrn if you prefer an even more user-friendly approach to the data!
